ProjectLab
What is the ProjectLab??
The Biology & Biotechnology ProjectLab provides MQP opportunities for students to work on projects directly affiliated with department faculty research in the in the department’s MQP lab on campus. This lab is located in Goddard Hall (GH 006) along with the rest of the teaching labs. This year 10 projects are potentially available in the project lab that accommodate multiple student teams and approaches, and range from animal to plants in the areas of study. Each project is proposed and sponsored by a Biology & Biotechnology faculty member and designed so that the data from the project will contribute directly to that faculty member’s current research. Daily work on the project will be overseen by the Sponsoring Faculty member, Project Lab Director (Mike Buckholt) or other biology faculty in Goddard Hall. Weekly meetings of the teams, the Goddard Hall faculty advisor, and the faculty sponsor will allow students to present their work to the group and to understand how their findings, along with those of other students working on the project, contribute to the overall research goals. Projects will run throughout the academic year and may start or continue into E term if student interest warrants. Not all students need to be registered for every term. Varying schedules of terms and credits can be accommodated.
Students interested in working in the ProjectLab should apply using the attached form or the form at https://web.wpi.edu/Images/CMS/Bio/new-projectlab-application.pdf
and return them to Mike Buckholt at mbuckhol@wpi.edu or just send me the info if you have problems with the form.
The Projects
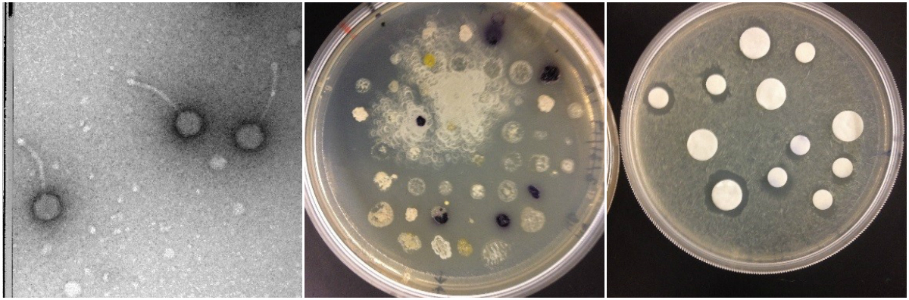
1&2 Antibiotic and Bacteriophage discovery Projects
Antibiotic resistance is a growing problem. Recently there have been too few new drugs or ways of combating infection entering the drug pipeline. This project will allow students to choose from a range of options to search for solutions to this problem. Because WPI is part of national effort to find antibiotic producing organisms from environmental samples (BB 2905/290X/2915) Microbes to Molecules) students can opt to test putative antibiotic producing samples already isolated and work on purifying and characterizing the antibiotic compounds they produce to determine if they are new and novel. Students could also opt to start from scratch in the isolation process or even explore what types of environmental samples yield the best results. If you are more interested in a molecular approach the project can use transposon inactivation of the antibiotic activity of a putative isolate to try to isolate the genes involved in the antibiotic activity.
Students may also choose to explore the option of isolating bacteriophage that might have therapeutic benefits. Students can opt to continue work from the two PHAGE HUNTERS course (the hunt and the analysis) using wet lab techniques, bioinformatics techniques or a combination of the two. Students can even isolate a new phage from the environment and try out new host bacteria. The direction of these projects can be very student driven and they may be of interest to students that have taken Microbes to molecules or Phage Hunters but taking the course is not a requirement.
Techniques: Will vary depending on the particular project but may include aseptic technique, inhibition assays, TLC, extractions, gel electrophoresis, bioinformatics, phage and bacterial isolation, restriction digestion, PCR, DNA sequencing, transduction, transposon inactivation and HPLC
Preferred team size 2-4 students
Project code: MB2-MM19 or SP19 advisors Mike Buckholt and JoAnn Whitefleet–Smith Depending on project
Extending Microbes to Molecules or Phage Hunters
3. Complementary and Alternative Medicine: Assessing the role of Phytoestrogens in breast cancer
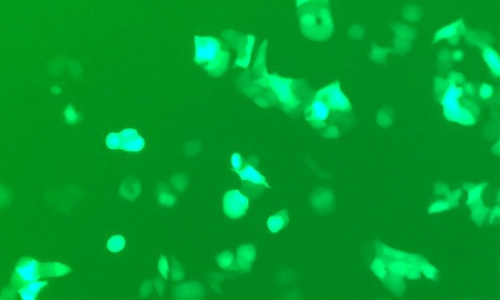
Phytoestrogens are plant compounds that can mimic the effects of human steroids. Traditional hormone replacement therapies have been associated with several health concerns, including a potential increase in the risk of breast cancer. Phytoestrogen products are often marketed as a safe and natural dietary alternative to estrogen and marketing claims include mood stabilization and an increase in overall health and wellness. However, these products, which are available over-the counter, vary considerably and are not well characterized or tested. Additionally, the dose in supplements is often far higher than those in dietary sources.
Using a panel of well characterized estrogen responsive breast cancer cell lines, we can test the effects of available phytoestrogen products and their individual components on cell growth. Preliminary studies suggest that some of these compounds may actually have anti-proliferative or pro-apoptotic effects, which may make them attractive from a therapeutic standpoint. Our cell model system allows us visually assess binding to the estrogen receptor and to regulate hormone receptor levels so we can begin to dissect the specific action of these products.
Techniques: Cell culture, Microscopy, Plate assays, Protein assays, Immunoblotting, Protein gels, HPLC
Preferred team size 1-3 students
Project codes: MB2-PE19, Advisors Mike Buckholt and Jill Rulfs
4. Artemisia annua: does the plant contain anti-cancer compounds
Artemisinin is a compound derived from the plant Artemisia annua, that has been used as a treatment for malaria. There is also some evidence that it can affect the growth of cancer cells. Recent work has suggested that the plant itself can be used as a therapeutic for malaria without the added step of artemisinin extraction. However, the plant contains a number of compounds which have the potential to also affect cell growth and proliferation. This project aims to further assess the anti-cancer claims using whole plant extracts and cancer cell lines to evaluate whole plant extracts as a natural anticancer therapies.
This is a new project for the project lab and will allow for a lot student input and direction.
Techniques: Cell culture, Microscopy, Plate assays, Protein assays, Immunoblotting, Protein gels, HPLC
Preferred team size 1-3 students
Project codes: MB2-AR19, Advisors Mike Buckholt and Jill Rulfs
5. Roundworm/nematode diversity in Central Massachusetts
Did you know that roundworms are one of the largest groups of animals inhabiting the planet? These organisms are present on land and water. There are both benign and pathogenic species found in many locales. Very little is known about the various species that inhabit the soil of New England and Massachusetts in particular. Knowing what species can be found here is the first step in understanding more about our soil ecology and the distribution of roundworms. The purpose of this MQP is to begin defining the species that can be found in our area. The initial phase of the project will involve collecting diverse soil samples from the surrounding area. The next phase of the project will include isolating and characterizing the nematodes present in the soil samples on both the macroscopic and molecular levels. These investigations will include characterization of brood size, morphometry and PCR techniques to complete the classification of nematodes. The data generated from the experiments will be then be overlaid on to the map of Central Massachusetts to show the species distribution. A secondary goal of this project is to develop a set of techniques that can eventually be used to turn this into a citizen science project in which K-12 students from all over could participate.
Here is the example of one such nematode collected from California.
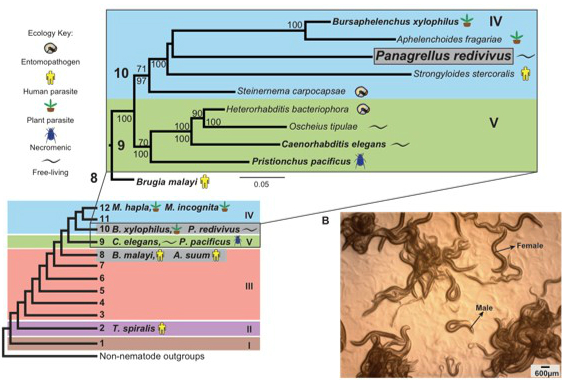
Project Code: MB2-RW17, Advisors Mike Buckholt and Jagan Srinivasan
Techniques: microscopy, morphometry, PCR, Bioinformatics
6. Stressed out! Environmental toxins and the cellular stress response
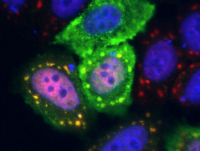
The cellular stress response protects the cells of our bodies from harm during periods of acute stress such as abrupt temperature changes, energy depletion, and brief high-dose exposure to toxins. While we typically think of exposure to toxic or stressful conditions to be a bad thing, many studies have shown that low levels of chronic stress can actually be beneficial to cellular survival when cells are later exposed to an acute stress, a phenomenon known as adaptive stress response or hormesis. In this MQP, students will study the effects of a variety of environmental contaminants on the acute and adaptive stress responses, and on cellular survival during stress. Common contaminants to be investigated include heavy metals (arsenic, lead), plastic components (bisphenols, phthalates), cosmetic additives (parabens) and pesticides (2,4-D, glyphosate, and others). The specific stress conditions or toxins investigated can be selected based on students’ interests. This project
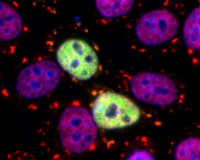
utilizes a well-characterized assay system and has been very successful in collecting data. Publication of previous MQP studies on bisphenols is currently in progress, and other publication opportunities are possible for motivated students. Students will learn mammalian cell culture, fluorescence microscopy, digital image analysis, Western blotting, and data analysis.
Project Code: NF-AS19, Advisor Natalie Farny
Techniques: cell culture, fluorescence microscopy, digital image analysis, PCR, Western blotting, and DNA cloning and expression.
Students: 2-6 (two teams possible)
7. Synthetic Biology
Are you interested in genetic engineering? Would you like to try and solve a global problem using biology as a tool? This MQP opportunity will give students experience in the emerging field of Synthetic Biology, defined loosely as 1) the design and construction of new biological parts, devices and systems; and 2) the re-design of existing natural biological systems for useful purposes. Specific topics for these projects will be student-defined and will begin with the framing of a problem. Students will then use the scientific literature to design solutions to that problem, which they will then build and test in the lab. The model organism we will use for this project is the bacteria E. coli. You will be amazed what these little bacteria can do! Interested students are encouraged to visit igem.org/About to learn more about synthetic biology and view sample projects.
Project Code: NF-SB19, Advisor Natalie Farny
Techniques: cloning, transformation, bacterial culture, PCR, sequencing, microscopy, plate reader assays
Preferred team size: 2-3 students
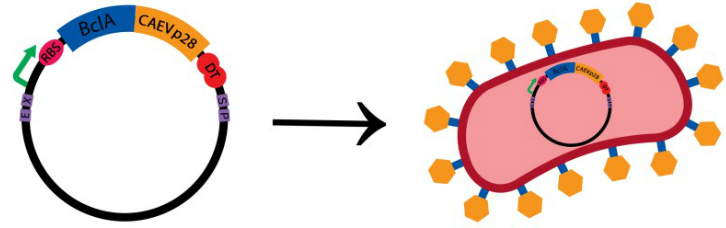
8. Mycobacterial Gene Regulation and Survival of Stress
The project team will embark on a mission to understand how the bacterium that causes tuberculosis regulates gene expression in response to stress. Students will directly contribute to both the research mission of Prof. Scarlet Shell's laboratory, as well as the BB 3527 lab course. Our goal is to discover how gene regulation mediates response to environmental triggers in Mycobacterium smegmatis, a soil bacterium that serves as a realistic and safe model for M. tuberculosis. Specifically, students will determine how upstream untranslated regions (UTRs) promote bacterial cell survival by affecting mRNA stability and/or translational efficiency of stress-responsive genes. Students will perform DNA assemblies, quantitative PCR, and fluorescence microscopy to design, create, and evaluate reporter genes to assess their

UTR. By gaining a better grasp on the molecular basis of how M. smegmatis responds to its environment and withstands stress, we may identify targets for therapies that better fight the bacterium responsible for the devastating human disease tuberculosis.
Project Code: LR-SM19, Advisors Lou Roberts and Scarlett Shell
Techniques: cloning, transformation, bacterial culture, PCR, sequencing, DNA assemblies, quantitative PCR, and fluorescence microscopy
2-4 students
9. Evolution of Social Behaviour

“Sociobiology” is a field of biology that seeks to understand the evolution of social organization, which is mediated by a collection of behaviors that allow individuals to navigate an environment of conspecifics in a way that facilitates their own survival and fitness. Species vary in the complexity and extent of their social organization. For example, organisms that live in dense populations are under strong evolutionary pressure to use social strategies that result in the best outcome in terms of individual fitness. Crayfish are often used as models for the study of social behavior because
they tend to live in dense aggregations and have consequently evolved a complex set of social behaviors. For example, crayfish establish clear hierarchies of social dominance that shape the roles that individual crayfish play in a local population and that determine allocation of resources like food, shelter, and mates. This project will use crayfish as a model to test hypotheses about the role of environmental and ecological factors in the evolution of social behavior, to address questions such as: How do individual crayfish communicate dominance status to other crayfish, and how do individuals process this information to make beneficial decisions about whether to fight or back down? What traits do individuals seek in potential mates, and why? How does the presence of other species, such as competitors or predators, affect social interactions among crayfish? Experiments are likely to be laboratory-based, though, depending on student interest, there is the possibility of field experiments.
This project or parts of it can be started in E term.
Techniques: field collection, behavioral observation, animal handling, statistical analysis, experimental design.
Number of students: 1-4
Project Code: LM-CB13
10. Environmental Impact of Invasive species
Invasive species (species introduced outside their natural ranges by human activities) are thought to be important drivers of biodiversity loss and may result in changes to environments at both

local and regional scales. In North America, freshwater faunas are particularly vulnerable to ecological changes, because of heavy manipulation of habitats by human activity. In addition, North America harbors a substantial majority of the world's biodiversity in freshwater crayfish, many of which are considered to be species of conservation concern. Interactions with invasive crayfish species are known to cause ecological changes in many parts of the world, especially in North America and Europe. This project will use a known invasive crayfish species, Procambrus clarkii, as a model to examine the effects of invasive species on local ponds and streams. Depending on student preferences and the terms this project runs, experiments will involve anything from setting up outdoor enclosures to individual microhabitats in the lab. Possible studies will involve water and sediment testing for various nutrients, flora, and fauna and may change depending on the interest of the student(s).
This project or parts of it can be started in E term.
Techniques: field collection, observation, water testing, animal handling, statistical analysis, experimental design.
Number of students: 1-4
Project Code: LM-CB14