Systems and Theoretical Biology
In systems biology, we use mathematical tools to identify the basic principles that underlie complex biological systems. We aim to develop quantitative and predictive descriptions of these systems in health and disease.
At WPI, Systems and Theoretical Biology is a research focus area, with faculty from the Mathematical Sciences, Physics, and Chemistry & Biochemistry Departments. See below for details from individual faculty.
Look for relevant talks to be posted here.
Contact Sam Walcott (swalcott@wpi.edu) for questions/comments about Systems and Theoretical Biology at WPI.

Sam Walcott (Mathematical Sciences)
I develop mathematical models of biological systems at the molecular scale to understand macroscopic biological function. Past questions of interest include: How do a molecule's mechanical properties influence the behavior of a cell? How do single molecule measurements relate to muscle function? To answer such questions, I use a combination of computer simulations and mathematical analysis. This theoretical work is complemented by experiments performed by my collaborators.
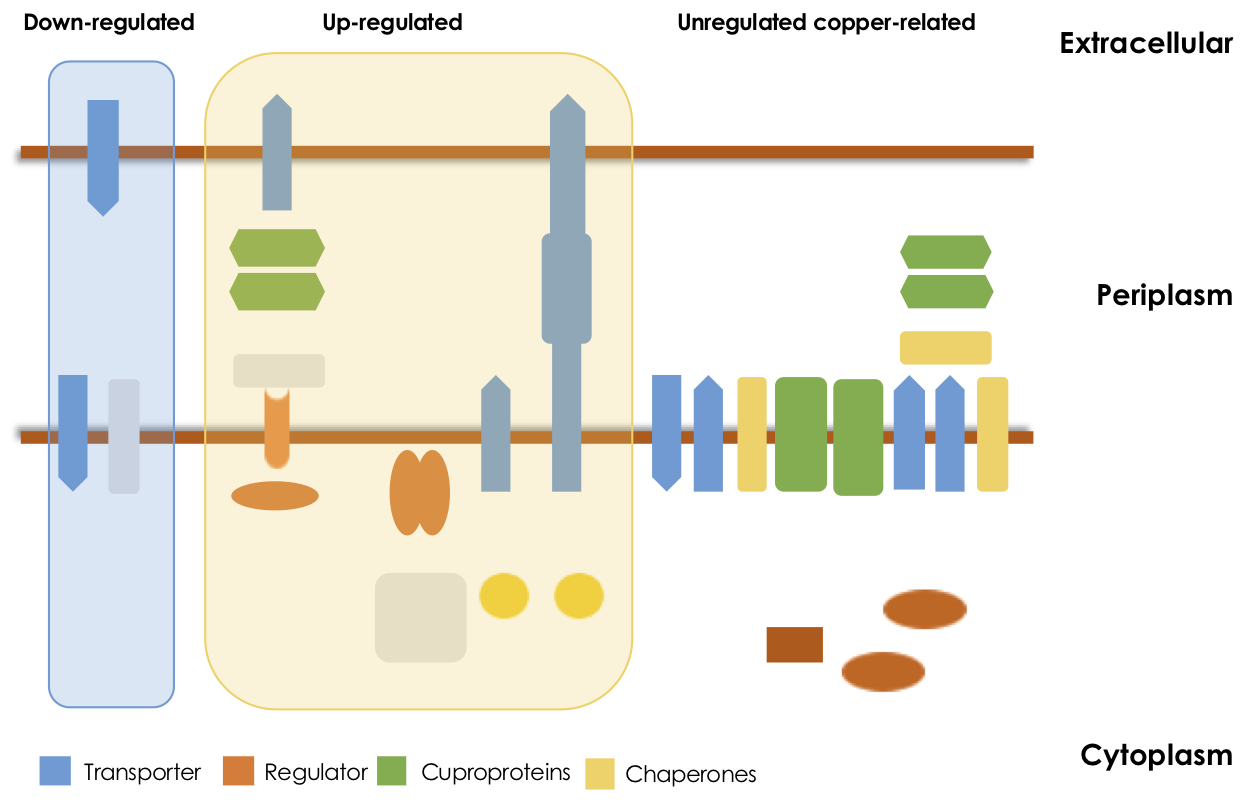
José Arguello (Chemistry and Biochemistry)
Our group is interested in understanding transition metals homeostasis in pathogenic bacteria (Pseudomonas aeruginosa and Salmonella). We aim to define and model cellular metal distribution networks by using system biology approaches, transcriptomics and metalloproteomics among others. In addition, we are integrating Cu+ distribution fluxes into mechanistically driven mathematical models. Departing from reductionist approaches, the project shifts the analysis of heavy metal homeostasis by considering the full range of involved elements, the biochemical equilibria in which they participate, and the integrated system response to environmental challenges.
Andrea Arnold (Mathematical Sciences)
Andrea Arnold: My research addresses solving inverse problems and quantifying uncertainties in biological and medical applications. This work involves developing and utilizing sophisticated computational mathematics and statistical techniques to estimate unknown quantities of interest from real-world data, which is often obtained experimentally. Recent projects include mathematical modeling and simulation for problems in neuroscience, epidemiology, and medical laser surgery. Learn more about Andrea Arnold.

Germano Iannacchione (Physics)
I am an experimentalist focusing on thermo-physical properties of soft-matter that includes biomaterials. A recent set of projects involved the study of the drying of bio-colloidal drops (various protein solutions, mixtures including liquid crystals, whole blood, etc.) as a function of concentration monitoring the drying evolution and final structure/morphology of the dried film. This research fits into the general area of emergence in non-equilibrium systems. Learn more about Germano Innacchione.
George Kaminski (Chemistry and Biochemistry)
My research is focused on building bridges between mathematical models of biophysical systems and practically interesting applications of these models. The main goal is to find balance that makes the calculations both simple and practical, and, at the same time, sufficiently accurate and robust. We use computer molecular simulations in my group, and the main direction is currently in modeling proteins and protein-metal complexes. Learn more about George Kaminski.
Adam Lammert (Biomedical Engineering)
My research centers on the mechanisms of perception and action in biological systems. I work on (a) modeling how the nervous system coordinates complex movements like those observed during locomotion, dexterous manipulation and speech production, and also (b) quantifying variability in such movements across contexts and conditions, including those associated with pathology. These efforts incorporate elements from dynamical systems theory, control theory, signal processing and statistical modeling.
Sarah Olson (Mathematical Sciences)
My group develops biophysical models that couple multiple scales with the goal of understanding the underlying biological processes and the emergent properties of these complex systems. Currently, our group uses sophisticated mathematical analysis and develops new computational methods to answer questions related to microorganism motility, cell division, tissue engineering, and cancer therapeutics. Learn more about Sarah Olson.
Suzanne Scarlata (Chemistry & Biochemistry)
Our lab uses biophysical approaches to understand the communication network that becomes active when extracellular agents interact with cells. Our recent work focuses on the series of concerted reactions invoked by neurotransmitters that allow neuronal connections to form and break during processes such as learning and memory. We have recently delineated how neurotransmitters induce chemical signals to produce mechanical changes to alter cell connections. Although these signaling networks are critical in neuronal cells, they are also important in muscle and other cell types. Scarlata Lab.
Kun-Ta Wu (Physics)
My group works on self-organization of active fluids which are composed of entities that consume fuel and propel themselves. This feature of active fluids allows for accomplishing tasks beyond the limits of conventional passive fluids such as transporting cargo without external pumps. While there are various types of active fluid such as bacterial suspension and self-propelling colloidal systems, my group focuses on kinesin-driven, microtubule-based active fluid, which hydrolyzes adenosine triphosphate to create chaotic flows at low Reynolds number systems. In this kinesin-microtubule system, my group asks miscellaneous fluid-related questions such as how active fluid can create active turbulence to enhance mixing efficiency in low Reynolds number systems where conventional high Reynolds number turbulence is suppressed and how we can regulate the chaotic flow of active fluid to a river-like coherent flow that has potential to create an active fluid-powered machine. Learn more about Kun-Ta Wu.
Min Wu (Mathematical Sciences)
Min Wu: My research centers on developing mathematical models and numerical methods to understand growth and morphogenesis in living systems. I currently work with plant-cell biologists to understand the control(s) of walled-cell development and morphological variations and bioengineers to understand and guide large-scale soft tissue migrations. Learn more about Min Wu.









