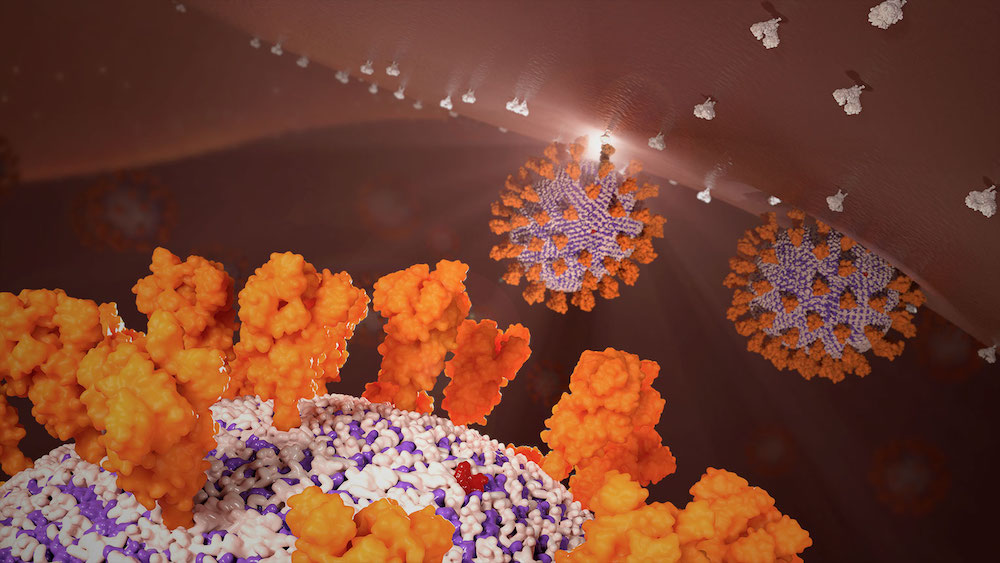
A Worcester Polytechnic Institute (WPI) bioinformatics researcher has created and unveiled a structural 3D roadmap of the new coronavirus (2019-nCoV), a major development that potentially holds the key to understanding the spread and treatment of the deadly virus.
Using a recently published viral genome of the Wuhan coronavirus made available on the National Center for Biotechnology Information website, Dmitry Korkin and a team of graduate students used molecular modeling to reconstruct the 3D structure of major viral proteins and their interactions with human proteins.
“We’re confident that our data and visual models could provide the guidance for experimental scientists worldwide who are working feverishly to solve this pandemic,” said Korkin, associate professor of computer science at WPI and director of the university’s bioinformatics and computational biology program.
In effect, Korkin said, he and his team have created a structural genomics map of the coronavirus that will be made available to researchers and anyone worldwide.
“Anyone can download our models, get any kind of information and use it for research,” he said. “This is one of the first examples of how data-driven science can quickly respond to this challenge.”